Quick start guide
See vignette("authentication") for instructions on
creating your personal API token for accessing data. To begin using
pluto, load the package and log in with your personal API
token:
library(pluto)
pluto_login("<YOUR API TOKEN>")Interacting with Pluto data models
Pluto data is stored in several data models, which are described in
depth in vignette("pluto_models"). For the purposes of this
quick start guide, we’ll highlight the main ones:
-
Project- collection ofExperiments -
Experiment- a data set containing-
type- a known/predefined assay type in Pluto (e.g.rnaseq,cutandrun,proteomics) -
Sample Data- tabular annotations and other metadata associated with samples -
Assay Data- tabular measurements from the assay (e.g. a counts matrix for gene expression experiments) -
Analysis- known/predefined analyses that can be run in Pluto (e.g.differential_expression,umap)-
Results- tabular results generated by anAnalysis, format depends on the kind of analysis being run
-
-
Using these models, you can read data into your Pluto R scripts in a flexible way to serve a wide variety of needs and applications. The basic examples below are intended to illustrate some foundational building blocks when working with Pluto data models.
Fetch data for a single experiment in Pluto
For this example, we’ll use a public ChIP-seq experiment from GEO
(GSE150555) that was imported to Pluto:
Effect
of WDR5 degrader on H3K4me2 in human cancer cell lines. The Pluto ID
for this experiment (found in the url) is PLX191681.
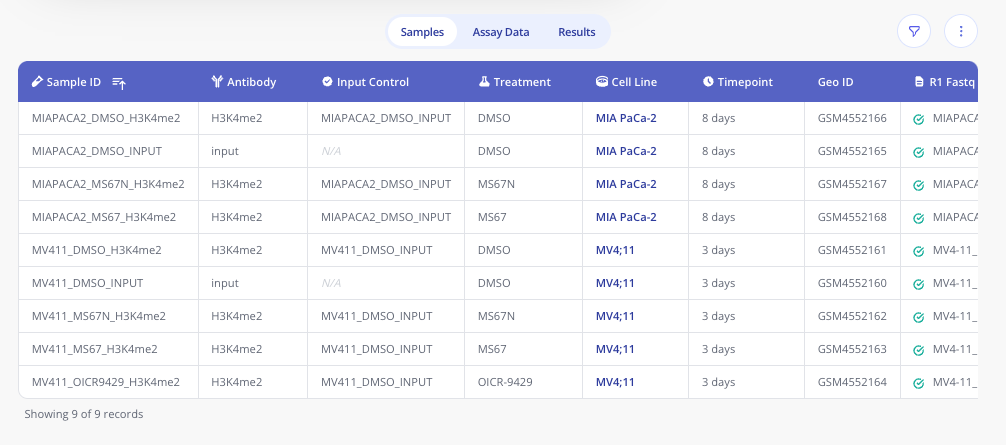
Sample data
In Pluto, the sample annotations are stored in a tabular format with automatic enriching of metadata for fields like cell lines.
To read these sample annotations into your R script, provide the
experiment ID to the pluto_read_sample_data() function:
experiment_id <- "PLX140206"
sample_data <- pluto_read_sample_data(experiment_id)